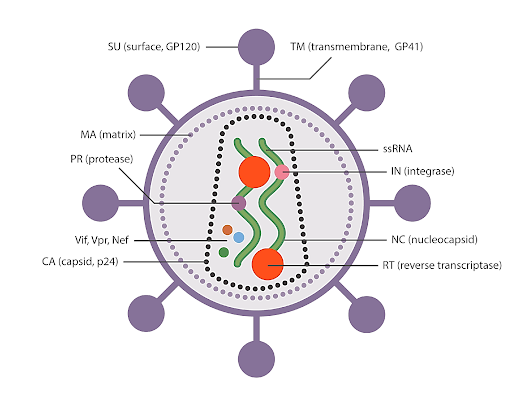
Omicron Variant - A Threat to Mankind
The Omicron variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was reported as a novel variant to the World Health Organization (WHO) on November 24, 2021. It is also known as B.1.1.529, as a variant of concern (VOC) on November 26, 2021. The Omicron variant's genome differs from other known variants in several ways. The Omicron variant's emergence has been attributed to three distinct processes: silent evolution in a population with limited sequencing; long-term evolution in one or a small number of people with chronic infection or evolution in other animals, particularly rodents. It is noteworthy that the Omicron variation has evolved into five lineages known as BA.1, BA.2, BA.3, BA.4 and BA.5. However, BA.2 is increasingly displacing BA.1 in a number of nations, including Denmark, Nepal, and the Philippines. BA.1 was previously the most extensively distributed strain around the globe. Only a few hundred instances, at most, of BA.3 have been reported to be transmissible. Due to their infectious and vaccine-escape alterations, the Omicron variants have created widespread anxiety and alarm. Currently, up to 60 mutations in the BA.1 lineage have been found; of these, up to 38 have been found in the spike (S) protein, one in the envelope (E) protein, two in the membrane (M) protein, and six in the nucleocapsid (N) protein. The BA.2 lineage has 57 mutations, 31 of which are in the S protein, whose N-terminus differs greatly from that of BA.1. The S protein's receptor-binding domain (RBD) binds to the host angiotensin-converting enzyme 2 (ACE2) receptor and has the ability to boost infectiousness and facilitate escape from vaccine-induced neutralising antibodies.
The RBD of the S protein mutations has therefore garnered a lot of research interest. Twelve mutations, including G339D, S373P, S375F, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, and Y505H, are shared by BA.1 and BA.2 in the RBD. Only BA.1 was associated with S371L, G446S, and G496S, while BA.1.1 was the only member of this group to include R346K. BA.2 shares T376A and D405N with BA.3 and has two distinct RBD mutations, S371F. Some of these changes, which are known to improve transmissibility, increase viral binding affinity, and cause antibody escape, have also been discovered in previous forms. For instance, it has been proposed that mutations in the residues K417, E484, and N501, which have previously been identified in Beta (B.1.351) and Gamma (P.1) variations, may mediate escape from vaccine-induced neutralisation. Most of the remaining Omicron mutations have unknown effects, so it is still uncertain how the virus behaves and how susceptible it is to inborn and vaccine-mediated protection. Additionally, this novel variation can re-infect those who have already contracted other SARS-CoV-2 forms. According to a recent study, people who are recuperating from illnesses caused by previously common variants are likely to contract the Omicron variant. This study demonstrates that Omicron mutations circumvent the immunity brought up by the earlier infection.
There is an urgent need for in-depth research and thorough knowledge of Omicron as it poses a major risk to public health and may jeopardise efforts to contain the COVID-19 pandemic. Recent years have seen a number of developments in our understanding of the Omicron variants.
Transmissibility and Infectivity
SARS-CoV-2 enters the host cell through membrane fusion with the aid of furin and type II transmembrane serine protease (TMPRSS2) or cathepsin L,12 which is a vital step in the infection process. S protein is used to bind to the primary receptor ACE2 on the host-cell surface. The Omicron variation is more contagious than earlier versions but shares a similar infectious mechanism. The results of preliminary research on Omicron's contagiousness and transmission are discussed below.
ACE2 host cell receptor binding
ACE2 is a major receptor of SARS-CoV-2. The ACE2 binding affinity for the S proteins of Omicron is the key factor determining the infectiousness of a virus. One of the key elements affecting the infectiousness of a virus is the binding affinity of ACE2 for the S proteins of Omicron. The reports on the Omicron variant's binding affinity to ACE2 to date have been inconsistent, probably as a result of variations in experimental studies or protocols. Numerous investigations have revealed that Omicron RBD has a 1.5–2.8 times greater binding affinity to ACE2 than the wild-type (WT). The Delta variant, Omicron RBD, exhibits a similar or weaker binding ability to ACE2 than the formerly prevalent Delta variant. Overall, the Omicron RBD mutation did not hamper its ability for receptor recognition and binding to ACE2, and the Omicron RBD can effectively bind to human ACE2 receptors for its host-cell entry.
Host-cell entry
The S protein mediates SARS-CoV-2 entrance into host cells once it binds to host receptors. The S1 and S2 subunits make up the S protein. The transmembrane region of the S protein, which anchors the S protein to the cell membrane and facilitates fusion of the viral membrane with cellular membranes, is found in the S2 subunit, while the RBD, which binds to ACE2, exists in the S1 subunit. For a virus to enter host cells, the S protein must be cleaved at the S1-S2 and S2 sites. This cleavage is carried out by furin, type II transmembrane serine protease (TMPRSS2), or cathepsin L26. There are two different SARS-CoV-2 entrance routes that are mediated by TMPRSS2 and cathepsin L cleavage at the S2 site. Six distinct mutations on S2 (N764K, D796Y, N856K, Q954H, N969K, and L981F) seen in the Omicron variation have never been found in any other VOC. According to recent research, the Omicron spike pseudotyped virus preferred the endosomal entry pathway over the plasma membrane entry route, and infection was reduced in TMPRSS2 expressing cells but enhanced in cells that support endosomal entrance. These findings demonstrate why Omicron replicates more quickly in the upper airways than in the lungs compared to other variants, and they also suggest that mutations on the Omicron S protein (non-RBD) may alter the route of viral entry into host cells. This is associated with a shift in cellular tropism away from TMPRSS2-expressing cells.
The furin cleavage site area of the Omicron variant also carries three alterations (P681H, H655Y, and N679K). It has been shown that the basic mutation P681H in the polybasic cleavage site (PBCS), which is also present in Alpha and Gamma, facilitates furin-mediated cleavage of the S protein and may therefore increase infectivity. However, among the SARS-CoV-2 variants, Omicron's amount of cleavage by furin is the lowest, suggesting that other alterations close to the furin cleavage site may seriously impair its cleavage. Additionally, the Omicron variant's fusion ability is the slowest of all SARS-CoV-2 variants and is comparable to SARS-CoV-1 (Figure 1).
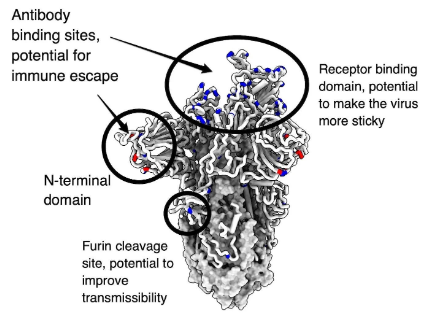
Fig 1: Detailed structure of Omicron variant.
Transmissibility
The high transmissibility of the Omicron variant is a major cause of global concern. Since the advent of Omicron, it has rapidly replaced Delta as the dominant strain worldwide. In the US, Delta accounted for 99% of new cases on December 4, 2021; however, Omicron accounted for more than 95% by January 8, 2022. The basic reproduction number (R0) of the Delta variant was between 3.2 and 8.36 The transmissibility of the Omicron variant is ~3.2 times that of Delta, and the doubling time is approximately three days. Generally, BA.2 is ~1.4 times more transmissible than BA.1, with a transmission rate of ~13.4% among household contacts, whereas that of BA.1 was 10.3%. The rapid spread of Omicron variant is due to its ability to evade the immune system and infect both previously healthy people and those who have received vaccinations. The binding affinity of Omicron RBD to ACE2 plays a vital role in transmission.
Pathogenicity
The Omicron variant's disease severity has generated a lot of debate and had a significant impact on public policies. According to mounting data, people with Omicron infections experience fewer symptoms than those with earlier variant infections. Additionally, the Omicron type is less likely to induce lung infection and more likely to affect the upper respiratory tract. The apparent decrease in pathogenicity is now, however, accentuated by improved immunity. According to UK research, three immunisation doses significantly reduced the likelihood of hospitalisation for Omicron compared to those who did not receive the vaccine. Similar defence may also come from prior infection. Omicron causes a significantly higher percentage of breakthrough infections than prior versions, which would lessen the severity we saw. Therefore, Therefore, the Omicron remains dangerous for those who are unvaccinated, especially the elderly. Omicron infection should receive significant treatment.
Immune Evasion
The Omicron variant's capability for immune evasion is a serious concern. It is highly transmissible due to its ability to bypass immune defences built up from vaccination or prior infection with another variant. According to UK data, BA.2 and BA.1 are both capable of evading the immune system. In addition, despite the modest number of cases that have been reported, a recent investigation demonstrated that BA.2 can re-infect BA.1 convalescent patients. The RBD region and N-terminal domain (NTD) of the Omicron variant have a number of alterations that are the primary targets of neutralization. Unprecedented complexities in mutation patterns can alter antigenicity and render immunity ineffective. The structure of Omicron's immune evasion mechanism is revealed using cryo-electron microscopy (cryo-EM). While a single-up conformation and an all-down conformation were shown in earlier variations, Omicron S-trimer only created one conformational state with one "up" RBD and two "down" RBDs. Mutations that disrupted antibody recognition resulted in steric conflicts, changed interactions at antibody-binding surfaces, and local alterations in the spike structure.
New Emerging Variant
The COVID variant omicron has rapidly grown into a number of subvariants since it first appeared in late 2021. An increase in COVID infections across China is being attributed to the subvariant, BF.7, which has recently been identified as the major variant spreading in Beijing. The omicron variant BA.7 has a sublineage of the omicron variant BA.5, abbreviated as BA.5.2.1.7. According to reports from China, BF.7 has the strongest capacity for infection among the other subvariants of Omicron. It spreads more quickly than other variants, has a shorter incubation period, and has a higher propensity to infect individuals who have already contracted COVID, received a COVID vaccination, or both. R0, or the fundamental reproduction number, for BF.7 is believed to range from 10 to 18.6. Accordingly, an infected person will spread the virus to 10 to 18.6 other people on average. Omicron has an average R0 of 5.08, according to the reported research. It is believed that the high rate of BF.7 transmission, along with the risk of masked spread brought on by the large number of asymptomatic carriers, is making it extremely difficult to contain the epidemic in China. Over time, new subvariants of omicron have emerged that are better able to bypass vaccination or past infection-induced immunity. BF.7 is no different. A particular mutation, R346T, exists in the SARS-CoV-2 spike protein in BF.7 (a protein on the surface of the virus that allows it to attach to and infect living cells). This mutation, which is also present in the "parent" form of BF.7, BA.5, has been associated with improving the virus' ability to evade neutralising antibodies produced by vaccination or prior infection. A recent study examined the neutralisation of BF.7 in sera (a component of blood that should contain antibodies) from triple-vaccinated healthcare workers, as well as patients infected during the omicron BA.1 and BA.5 waves of the pandemic. BF.7 was resistant to neutralisation, driven partly by the R346T mutation.
Vaccines
Since the COVID-19 pandemic outbreak, several vaccines have been developed to end it, although they haven't been as effective against the Omicron variety. The major vaccinations currently used worldwide are much less effective against Omicron. In more than 80% of blood samples from people who received two doses of the inactivated vaccines CoronaVac and BBIBP-CorV, the neutralisation activity against Omicron was below the lower limit of quantification. Additionally, the vectored vaccines Ad26.COV2.S, ChAdOx1-S, and Sputnik V did not successfully produce neutralising activity against the Omicron form. The most efficient RNA vaccines against the WT strain, such as BNT162b2 and mRNA-1273, were utterly ineffective against the Omicron variation in over 50% of individuals. Neutralizing antibody titers against the Omicron variant were significantly lower in serum samples from those who had received two doses of BNT162b2 or mRNA-1273 compared to the WT. Despite the fact that each study had its own unique samples and testing procedures, the results demonstrated that the neutralising potency of two doses of RNA vaccines against Omicron had significantly decreased. According to an observational study conducted in the United States, the projected effectiveness of two doses of RNA vaccines against Omicron was only 30% one month after the second dosage, with no effectiveness for BNT162b2 and mRNA-1273 three and six months, respectively, after the second dose.The clinical outcome of SARS-CoV-2 is significantly influenced by the cellular immune response. Multiple changes in the Omicron variant's spike protein let it evade antibody neutralisation, although T cells and other cellular immune response elements can still target Omicron and mitigate its negative effects. According to studies, the SARS-CoV-2 Omicron Spike exhibits cellular immunity that was previously acquired from infection or vaccination in a highly conserved manner. In previously infected or immunised people, the median relative frequencies of SARS-CoV-2 spike-specific CD4+ T cells and CD8+ T cells that cross-recognize Omicron are between 70 and 90 percent, yet there is still a 50% drop in 20% of people. Booster vaccination also improves T-cell reactions to Omicron spike.
Although standard vaccination doses are rarely successful in neutralising the Omicron variant, reported data suggest that booster vaccinations can more successfully establish neutralising immunity against Omicron. Following booster shots, neutralisation titers for Omicron had increased by a factor of twelve, showing a considerable decrease in resistance to the Omicron variant compared to the WT strain. Furthermore, compared to unvaccinated mice, the efficiency of the BNT162b2 and mRNA-1273 vaccines against Omicron rose to 65% and 72%, respectively, after three doses of administration. It's interesting to note that heterologous booster vaccination has the ability to provide sufficient neutralising efficacy, which is crucial for boosting the level of protection offered by some vaccines that produce insufficient neutralising antibodies. In contrast to homologous booster vaccination with CoronaVac, which served as an efficient substitute in response to the Omicron, heterologous booster vaccination with aerosolized Ad5-nCoV, a representative of adenovirus-vectored vaccines, produced greater neutralising antibody responses against the Omicron variant. The protective efficacy attained may depend on the vaccine type, although there is currently no proof that heterologous immunisation is superior to homologous vaccination. It is therefore required to conduct additional study on the mechanism and safety of heterologous vaccination, which could help address the issue of vaccine scarcity. Overall, routine vaccination doses are rarely sufficient to provide protection against the Omicron variant; as a result, homologous or heterologous booster vaccinations are required, which also emphasises the importance of an adequate supply and fair distribution of vaccines. Additionally, although cross-neutralization has been seen against previous variants, the plasma of recovering individuals can scarcely neutralise the Omicron variant. However, combining immunisation with infection can result in the production of superior neutralising antibodies. Given the high prevalence of infection, this group of people may just need a normal immunisation dose to obtain reliable Omicron protection.
Therapeutic-neutralizing antibodies
Therapeutic-neutralizing antibodies have a limited ability to neutralise Omicron. The majority of therapeutic neutralising antibodies that have received EUA authorisation or are in advanced clinical development stages have been unable to neutralise the Omicron variant. There are two categories of neutralising antibodies. The majority of neutralising antibodies belong to the first category, which includes the drugs LY-CoV555 (marketed as bamlanivimab), LY-CoV016 (marketed as etesevimab), REGN10987 (marketed as imdevimab), REGN10933 (marketed as casirivimab), COV2-2196 (marketed as tixagevimab), COV2-2130 (marketed as cilgavimab), and CT 16 Due to the loss of the antigenic epitope, the majority of these antibodies no longer neutralise Omicron. The primary places for the evasion are K417N, E484A, Q493R, and N501Y. The combination of COV2-2130 and COV2-2196 displayed neutralizing activity against Omicron variant; however, this activity is 12-200 times less potent than it is against the WT due to G446S and N440K mutations. However, it has been shown that this combination, particularly COV2-2130, continues to be active against BA.2, probably as a result of the lack of G446S. The second group is represented by VIR-7831/S309 (sold as sotrovimab), which competes with ACE2 seldom but identifies non-RBM epitopes shared by sarbecoviruses like SARS-CoV. S309 demonstrates resistance to Omicron's antibody escape, with just a two- to threefold reduction in Omicron's ability to be neutralised in comparison to the WT. Due to the proximity of the antigenic location, G339D and N440K are thought to interfere with the combination of S309 and spike protein.
The S371F mutation in BA.2 seems to have a larger detrimental effect on S309. Although BA.1 has the S371L mutation, BA.2 has the S371F mutation, which differs in its different resistance to neutralising antibodies. Brii-198 (also known as romlusevimab) is effective at neutralising Omicron, although its ability is a little less strong and is susceptible to inhibition by R346K. Additionally, S2X259 was less effective against the BA.2 strain despite having a significant neutralising effectiveness against the Omicron variety. The recently approved LY-CoV1404 (marketed as bebtelovimab) has a high neutralising capacity against all recognised variations. Except for N439 and N501, structural analysis showed that the LY-CoV1404 epitope was largely conserved. Fortunately, Omicron's N501Y mutation has no effect on how well it binds. Overall, the COVID-19 pandemic may be controlled to a large extent by neutralising antibodies that target the conserved epitopes of SARS-CoV-2 variants and other sarbecoviruses. SARS-CoV-2 is prone to mutation, creating neutralising antibodies that focus on relatively conserved regions is a useful strategy for fending off new variations like Omicron. The effectiveness of the aforementioned antibodies in treatment is still under question. Despite the small sample size, a randomised controlled experiment found that neither S309 nor Brii-198 exhibited efficacy in improving clinical outcomes. Moreover, clinical trials are still needed to confirm the effectiveness of LY-CoV1404.
Antiviral drugs
In addition to therapeutic neutralising antibodies and vaccinations, a number of antiviral medications and prospective therapies are being researched for use against Omicron. The Omicron form has been proven to be less responsive to antibody treatments; nevertheless, the response to antiviral medications appears to be different. Omicron is susceptible to the majority of antiviral medications under study, including remdesivir, molnupiravir, PF-07304814 (nirmatrelvir, a crucial component of paxlovid), EIDD-1931, ribavirin, favipravir, nafamostat, camostat, and aprotinin, according to in vitro testing. These medications have various modes of action, which suggests that the Omicron variant's drug sensitivity profile doesn't alter significantly in response to the medications. The replicase-transcriptase complex of the Omicron variety only has two missense mutations (nsp7-10, nsp12, and nsp14), which may not have much of an impact on the RNA-dependent RNA polymerase-inhibitor. However, current research is still at the cellular level, and more clinical results are needed to support these conclusions. In response to evolving viruses and the existing dearth of vaccinations and medications, it is vital to develop new broad-spectrum antiviral medications. Drugs that prevent viral reproduction and also prevent viruses from attaching to ACE2, are likely to be effective against the Omicron variant. A promising area of investigation is the ACE2-targeting antibody, which has been demonstrated to reduce the Omicron variation at the cellular level. Another strategy that might be investigated is enhancing the antibody responses biochemically.
Additionally, it has been noted that SARS-CoV2 viruses can be effectively captured by modified extracellular vesicles (EVs) loaded with palmitoylated ACE2 (PMACE2), which prevents the viruses from interacting with cell surface ACE2 and reduces infection both in vitro and in vivo. Furthermore, it has been shown that COVID-19 patients have reduced IFN responses, particularly in those with advanced disease, which could be a factor in the weak antiviral response. Targeting antiviral responses may therefore be crucial in controlling SARS-CoV-2. A recent study demonstrated that the SARS-COV-2 N protein suppresses the antiviral immune response by undergoing liquid-liquid phase separation (LLPS) with RNA, which is a critical part of antiviral innate immunity. It's interesting to note that none of the currently known SARS-CoV-2 variations, including Omicron, have significantly altered the N protein's 246-365 domain, which maintains the ability to separate phases. Therefore, focusing on the SARS2-N protein LLPS may be a promising treatment strategy to combat SARS-CoV-2 infection in WT and mutant forms.
To support the Indian scientific community and accelerate their research in SARS-CoV-2, we, BTL Biotechnolabs Pvt. Ltd., offer a wide range of SARS-Cov2 research products, including antibodies, Lentiviruses, specific recombinant proteins, peptide pools, pseudovirus neutralization assay kits, spike proteins, antigens, ELISA kits, rapid PCR kits, and many more.
We also provide custom services according to the experimental research design.
For more details, please follow the below link:
https://biotechnolabs.com
https://biotechnolabs.com/products
For product details, please connect with us at info@biotechnolabs.com.